AnVIL Community Conference 2024 Conference Report
November 12 - 13, 2024
Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
The 2024 AnVIL Community Conference united AnVIL program leads, developers, community members, and novice users for the very first time! The meeting featured presentations from invited speakers to share about their research done on AnVIL, hands-on Collaboration Fest (CoFest!) on analysis topics including polygenic risk scores and deep learning for regulatory genomics, and time for discussion with the full audience through breakout sessions and a town hall. Attendees joined in from across academic, government, and commercial organizations across the United States and Australia with 58 total attendees, 13 of whom attended virtually.

AnVIL Welcome and Roadmap
Michael Schatz, Johns Hopkins University, Baltimore, MD
Jonathan Lawson, Broad Institute of MIT and Harvard, Cambridge, MA
Robert Carroll, Vanderbilt University Medical Center, Nashville, TN
The AnVIL Community Conference opened with a welcome and presentation about the AnVIL project, plus a discussion of the roadmap for the next phase of AnVIL and the project components.
AnVIL Past, Present, and Future
Michael Schatz presented on the first five years of AnVIL, highlighting the major accomplishments in the phase of the project and discussed plans over the next five years. A key opportunity of AnVIL is the collaboration with all NHGRI genomic data-generating consortia. In the next five years, the group will work to enable scientific services that will 1) support analysis of the data with Machine Learning (ML) and Artificial Intelligence (AI) models; 2) enable data and metadata harmonization by improving variant discovery and interpretation with long reads, offering state-of-the-art references and optimized liftover and realignment methods, as well as partnering with NIH Cloud Platform Interoperability (NCPI) projects for remapping and harmonization; and 3) improve predictions of cloud costs with new cost estimation methods and tools for researchers.
Multi-Cloud Roadmap
Jonathan Lawson shared an update on Terra’s multi-cloud stance and what can be expected in the second phase of AnVIL. While Terra is pursuing a multi-cloud offering, the Terra analysis platform and the AnVIL corpus of data will continue to be available on the Google Cloud Platform. Terra is refocusing efforts to enable truly multi-cloud implementation by continuing work to enable Microsoft Azure with extended timelines while generalizing to also implement Terra on Amazon Web Services in the next 4 years. The team has already and will continue to make improvements for data submission processes and technologies. Additionally, the entire AnVIL Data Library is now available in the DUOS system, where researchers can submit data access requests for NHGRI DAC review. There is active policy work in process to enable automated data access requests for GRU/HMB consent codes. AnVIL can also support researchers responding to requirements set forth in the 2023 NIH Data Management and Sharing Policy.
Clinical and Phenotypic Data Roadmap
Robert Carroll highlighted the opportunities and plans for clinical and phenotypic data in the next 5 years of the project. Some initial work to create findability data elements has been done to help researchers understand the data that are available. The team will work to meet opportunities in AnVIL to improve view of metadata element availability and provenance, and offer an improved set of harmonized data with well-defined expectations on coverage and transformation provenance. This will include mapping to standard ontologies and supporting researchers to be forward-looking in creating data models intended for sharing and combining their project data. The vision is for the core AnVIL data model to pull in data with custom data models mapped to clinical or research standards and provide a set of tools that support the use of the clinical and phenotypic data available in AnVIL.
Invited Talks
The AnVIL Community Conference also hosted three talks from investigator teams using the AnVIL and other cloud resources to support their genomic analyses.
Unlocking the Power of Precision Genomics in the Cloud
Speaker: Tychele Turner, Ph.D., Washington University School of Medicine, Saint Louis, MO
Deciphering regulatory syntax and genetic variation with deep learning models
Speaker: Anshul Kundaje, Ph.D., Stanford University, Stanford, CA
Using AnVIL: Long-read sequencing for advancing rare disease diagnosis
Speaker: Benedict Paten, Ph.D., University of California Santa Cruz, Santa Cruz, CA
Breakout Sessions
The Breakout Sessions offered an opportunity for attendees to discuss topics of interest to the community. The two topics were a discussion of data and a discussion of tools, and had high engagement from users, developers, and program staff to discuss the opportunities and challenges in these areas.
Data
The key takeaways for the data breakout session were:
- It is of high value for researchers, reusability, and citability to have DOIs for datasets and potentially workspaces. Currently, users might mint DOIs elsewhere if they are expected/required. The NIH and DataCite Consortium have offerings that are fairly straightforward to use and set up with APIs to mint DOIs, which would be of interest to use for AnVIL.
- There are opportunities to make it easier to access non-AnVIL data, such as SRA / dbGaP, and participating platforms in the NIH Cloud Platform Interoperability (NCPI) effort. Research Authentication Service (RAS) will support easier access in the AnVIL to dbGaP as well (AnVIL is in the home stretch here!), as it can track granted dataset access across platforms. Global standards such as the GA4GH Data Repository Service (DRS) is also being adopted to support cross-cloud interoperability.
- Data harmonization in AnVIL is a major topic of interest. A number of metrics or summary information from the data available would be of high value to the community, such as depth of read coverage and other QC metrics for genomics data, as well as assay information, specimens, participants, and diagnoses. The community conveyed that if the team generating these metrics is trusted, it will be valuable to all researchers for data discovery, though some researchers may generate these metrics independently for specific analyses when necessary. Automation and centralization would generate consistent summary information and metrics, with review to ensure gaps are identified and addressed. The AnVIL team could assemble an initial list and seek community input.
- As researchers consider which resource into which to deposit their datasets, there can be some confusion on which repository is best suited. AnVIL is an NIH-designated data repository that is authorized to share controlled-access data derived from human samples, associated phenotypic data, and metadata with the research community in a manner that is consistent with the expectations put forth by the NIH Genomic Data Sharing (GDS) Policy. AnVIL has lots of documentation and the teams are available to support researchers through the data submission process.
Tools
The key takeaways for the tools breakout session were:
- How to convey results for analysts? While AnVIL is a robust data deposition and analysis platform, there are opportunities to develop and enable functionality for results report outs with visualizations. The best current example is Jupyter notebooks and also Shiny apps via RStudio, which is a common interactive results visualization tool. Enabling more developers to bring in their own visualization apps, potentially via Kubernetes helm charts, would support this well, acknowledging the security considerations that are required. seqr is an example of this.
- Multi-cloud analysis allows greater flexibility, but some challenges remain, for example, egress costs are not quite apparent on some platforms.
- There could be improved recommendations on how to access AnVIL-hosted datasets (DRS URIs) vs bring your own datasets. It would be helpful to provide guidance to researchers on how to best share their data with others more broadly through AnVIL.
- An area of focus with tool capabilities is to consider how to streamline onboarding and development for analysts who are very familiar with high-performance clusters (HPC). It will be important to support one-off analysis in one step when possible (e.g., VCF Tools already on HPC) and to strategize on how to better support HPC analysts bringing more complex or custom analyses to the cloud by:
- Breaking down the problem into individual steps (Dockstore tools),
- “A la carte” checkboxes for packages to install when launching an environment, or
- Lightweight Docker might help folks work more efficiently.
Town Hall
The Town Hall session was an open opportunity for a full audience discussion on any topics of interest to the community. The discussion spanned data management; data discovery; and AnVIL onboarding, features, and cost considerations.
The key takeaways from the town hall session were:
- AnVIL is evaluating long-term data management strategies around data storage, including autoclassing datasets with infrequent access, stances on data deletion, and sustainability.
- There is a need to advance computational tools for efficiency in analyzing large datasets in the cloud.
- There are opportunities to leverage new formats for storing large data, and a number of technologies are emerging that need evaluation.
- There are new technical capabilities with AnVIL to prevent redundant analysis such as aligning existing data to new references by effectively disseminating derivative data and making clear links to the original datasets. There are ongoing discussions for a new NIH policy to support the effective sharing of derivative data.
- With DUOS, AnVIL is considering ways to improve data discovery for researchers by packaging datasets for access (e.g., by datatype, similar processing, alignment to same reference).
- AnVIL sees opportunities to improve data discovery by supporting metadata search associated with controlled-access genomic dataset, since certain metadata types are not required to be controlled-access. A few proposals were discussed. AnVIL views it as critical to follow and respect data consent codes and policies around exposing data for search.
- AnVIL users described onboarding as successful with a project ready for testing, available cloud credits, ready capability of AnVIL to support GPU use and model customization, and quick and helpful troubleshooting with support teams. A limitation to onboarding includes costs of commercial cloud providers, even though some cost saving capabilities are possible to explore (e.g., spot instances).
CoFest!
CollaborationFests!, also known as CoFests!, are collaborative work events where we expand and improve the AnVIL community and the AnVIL ecosystem. At ACC2024, there were five CoFest! topics.
Deploying, Training, and Interpreting Deep Learning Models for regulatory genomics in AnVIL
Scientific Lead: Anshul Kundaje, Ph.D. & Vivekanandan Ramalingam, Ph.D. Stanford University Outreach Coordinator: Kate Isaac, Ph.D., Fred Hutch Cancer Center
The “Deploying, Training, and Interpreting Deep Learning Models for Regulatory Genomics in AnVIL” CoFests! track at the AnVIL Community Conference offered hands-on training for users and developers interested in applying deep learning to regulatory genomics. This track demonstrated how deep learning models can be utilized for functional genomics datasets, such as ChIP-seq and ATAC-seq, at scale via AnVIL. Participants developed a comprehensive understanding of the steps involved in deploying, training, and interpreting these models, including the available input options and how to leverage the resulting outputs to address various biological questions.
The key takeaways from this session were:
- Demonstrated deep learning models as WDL workflows to train at scale in AnVIL.
- Demonstrated interactive analysis with a custom environment within Jupyter Notebooks to interpret models.
- Going forward, the team will add more documentation to make the workspace more accessible to those who want to use the tool.
Polygenic Risk Score (PRS) Analysis in AnVIL
Scientific Lead: Matthew Lebo, Ph.D., Harvard Medical School
Outreach Coordinator: Elizabeth Humphries, Ph.D., Fred Hutch Cancer Center
This track, run by the AnVIL Clinical Resource team, consisted of both an overview and a hands-on workshop to provide individuals with an understanding of polygenic scores and how to run and evaluate them in AnVIL. First, we level-set by providing an overview of the current state of polygenic analysis, with a focus on polygenic risk scores (PRS). Next, we jointly worked with participants to run PRS analyses in AnVIL using the WDL framework. These tasks increased in complexity in terms of analytical components of the workflow, with the goal of enabling users to run the WDL on their own. We also engaged with participants to get feedback and create user-friendly documents to enable processing of this workflow once published to the broader community. Finally, we worked with more advanced users to generate a new WDL focused on the evaluation of PRS among a cohort of individuals.
The key takeaways from this session were:
- Learned about Polygenic Risk Scores (PRS) and Polygenic Score (PGS) Catalogue.
- Learned how to run a Polygenic Risk Score (PRS) WDL workflow on AnVIL.
- Visualized the data using RStudio.
- Lots of tracking and troubleshooting.
- Bonus: learned how to use Jupyter Notebook!
How to Run Your Tool in AnVIL with WDL
Scientific Lead: Allie Cliffe, Broad Institute of MIT and Harvard
Outreach Coordinator: Javier Carpinteyro-Ponce, Ph.D., Carnegie Institution
Workflow Description Language (WDL) not in your programming skillset? No problem! Participants learned to wrap their pipelining tool in WDL and run it in an AnVIL workspace in this CoFest! session. We went over WDL basics and how to run pipelines in AnVIL, then walked through how to run a participant’s own Unix/Python/R script. Along the way, they wrote a WDL and built a custom Docker to run it with. All experience levels were welcome and an outcome was to create resources (e.g., tutorials, AnVIL Book, or Cheat sheet) for others to use to run their non-WDL tool in AnVIL.

The key takeaways for this session were:
- Learned and discussed how to run workflows on AnVIL using WDLs, how to create a WDL locally wrapping a custom tool, upload to Dockstore, and import it into AnVIL.
- Discussed the option of testing WDLs locally, before importing it into AnVIL using miniWDLs.
- Created a Nextflow script, wrapped it into a WDL, imported it into AnVIL and successfully ran it!
Brainstorming Feature Requests for AnVIL
Scientific Lead: Ava Hoffman, Ph.D., Fred Hutch Cancer Center
The “Brainstorming Feature Requests” CoFest! track at the AnVIL Community Conference provided an opportunity for users and developers to directly communicate their needs to the AnVIL Outreach Team. Participants shared their experiences with the NHGRI Genomic Data Science Analysis, Visualization, and Informatics Lab-Space (AnVIL), highlighting both its strengths and areas for improvement.
The AnVIL Outreach team facilitated discussion, gaining insights into real-world use cases and challenges. This open dialogue helped identify gaps in functionality and potential enhancements that could significantly improve the platform’s utility for genomic researchers.
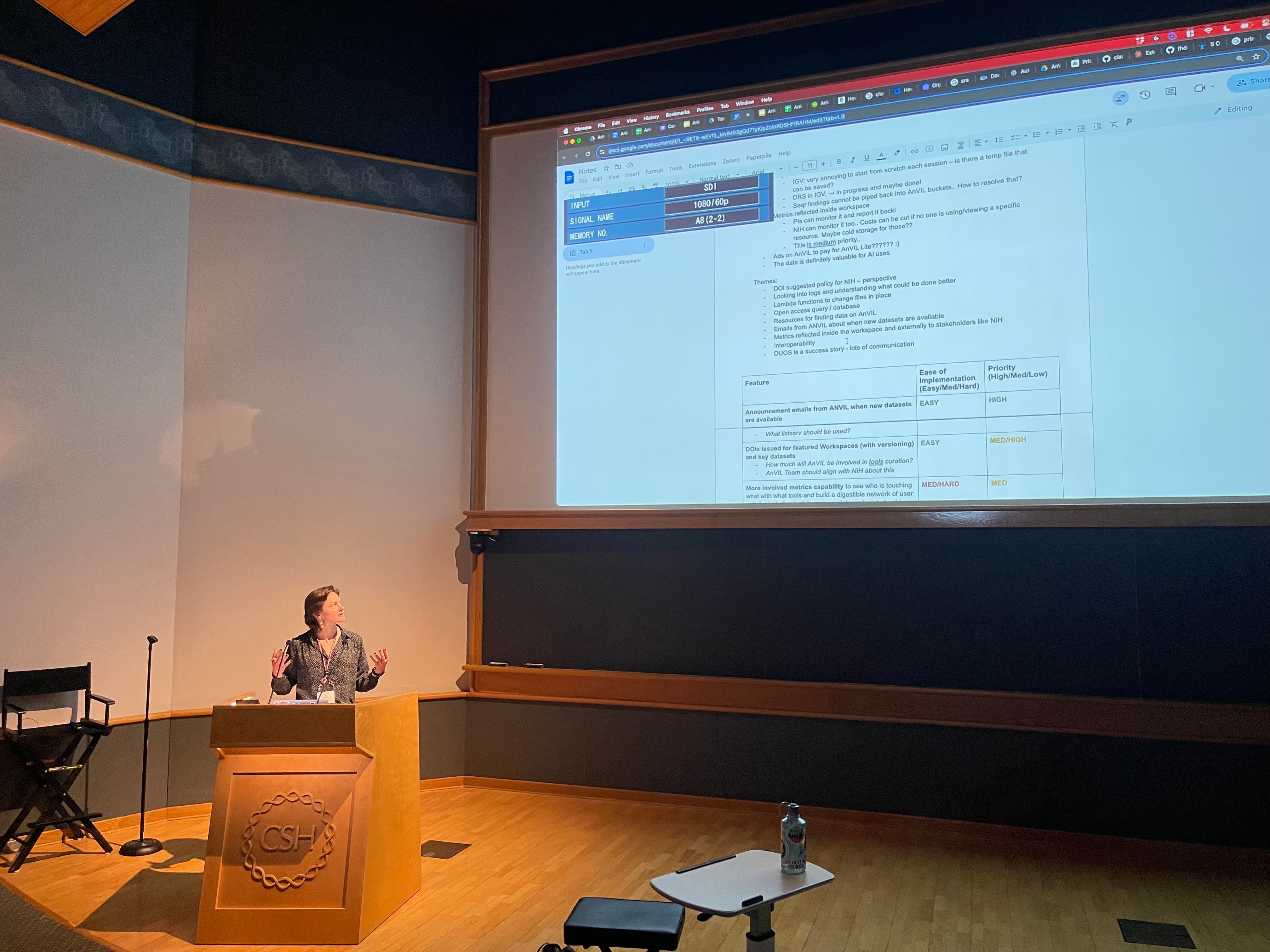
The key takeaways for this session were:
- Data Discovery and Communication
- Announcement emails can help advertise when new datasets are available.
- There are opportunities to develop better ways to find out about new datasets.
- Participants in this CoFest! expressed a desire for more clarity around AnVIL's unique offerings.
- Communication about the Terra Product roadmap and updates is an opportunity and is underway.
- Documentation and Metrics
- There is a need for better workspace metrics to track usage and activity.
- There is a need for PIs to have the ability to monitor usage and report back.
- There is interest in tracking both internal data (crash reports) and external data (page views, clones).
The CoFest! session created a table to summarize the features discussed, the ease of implementation, and the relative priority of each feature.
| Feature | Ease of Implementation (Easy, Medium, Hard, Underway, ?) | Priority (High, Medium, Low, Underway, ?) |
|---|---|---|
| Data announcement emails | Easy 🟢 | High 🟢 |
| DOIs issued for featured Workspaces (with versioning) and key datasets | Easy 🟢 | Medium 🟡 |
| Greater metrics capability | Hard 🔴 | Medium 🟡 |
| Lambda functionality to do small, serverless operations on files | ? | ? |
| Public URL to access non-controlled access data/workspace without a billing project | ? | ? |
| Terra product team roadmap users know what to expect | Underway 🟢 | Underway 🟢 |
| Clarity of AnVIL’s unique offerings | Medium 🟡 | High 🟢 |
| Run common tools across all the data at once | Hard 🔴 | ? |
AnVIL Developers Birds of a Feather
The AnVIL Developers Birds of a Feather session at CoFest! offered some open time for developers and community members to meet ad hoc to collaborate on cross-cutting topics to drive forward.
Summary
The first AnVIL Community Conference brought together the leads, developers, and users of AnVIL to share the exciting research performed on the AnVIL platform, key capabilities that are available now in AnVIL, and gather input on what offerings would best support the community for future use and further adoption of cloud computing for genomics research. Through keynote talks, hands-on workshops and collaboration sessions, and discussions in the breakout sessions and town hall, the conference highlighted opportunities and priorities for the AnVIL moving forward.
