Submission Process Overview
Overview
In order to submit data into AnVIL you will need to do the following:
- Register with dbGaP/Obtain required approvals.
- Set up your data model.
- Prepare your data for submission.
- Stage your data for ingestion.
- QC ingested data.
General Data Requirements
Make sure your data conforms to these overall data requirements, or contact the AnVIL data team.
Human Reference Genome
To maximize interoperability, all submitted human data should be based on the GRCh37 or GRCh38 human reference assemblies, though de novo assemblies and other references can be accepted if scientifically justified.
Register with appropriate NCBI resource (e.g. dbGaP, GEO)
Data in the AnVIL are stored in data workspaces. For controlled-access studies, consent codes from dbGaP are used to determine appropriate access to the data workspaces on AnVIL.
It is important that studies into the human genomic and phenotypic associations be registered with dbGaP so you can populate the data element dbGaP_study_ID (phsXXXXXX) in AnVIL. Additionally, NIH registration of studies provides AnVIL with information needed to set up the workspaces so that data can be appropriately organized by study (if there are multiple, e.g., within a consortia) and by consent group(s). Study registration often occurs at Just-in-Time (JIT) for NHGRI funded studies, and thus you may have already completed this step.
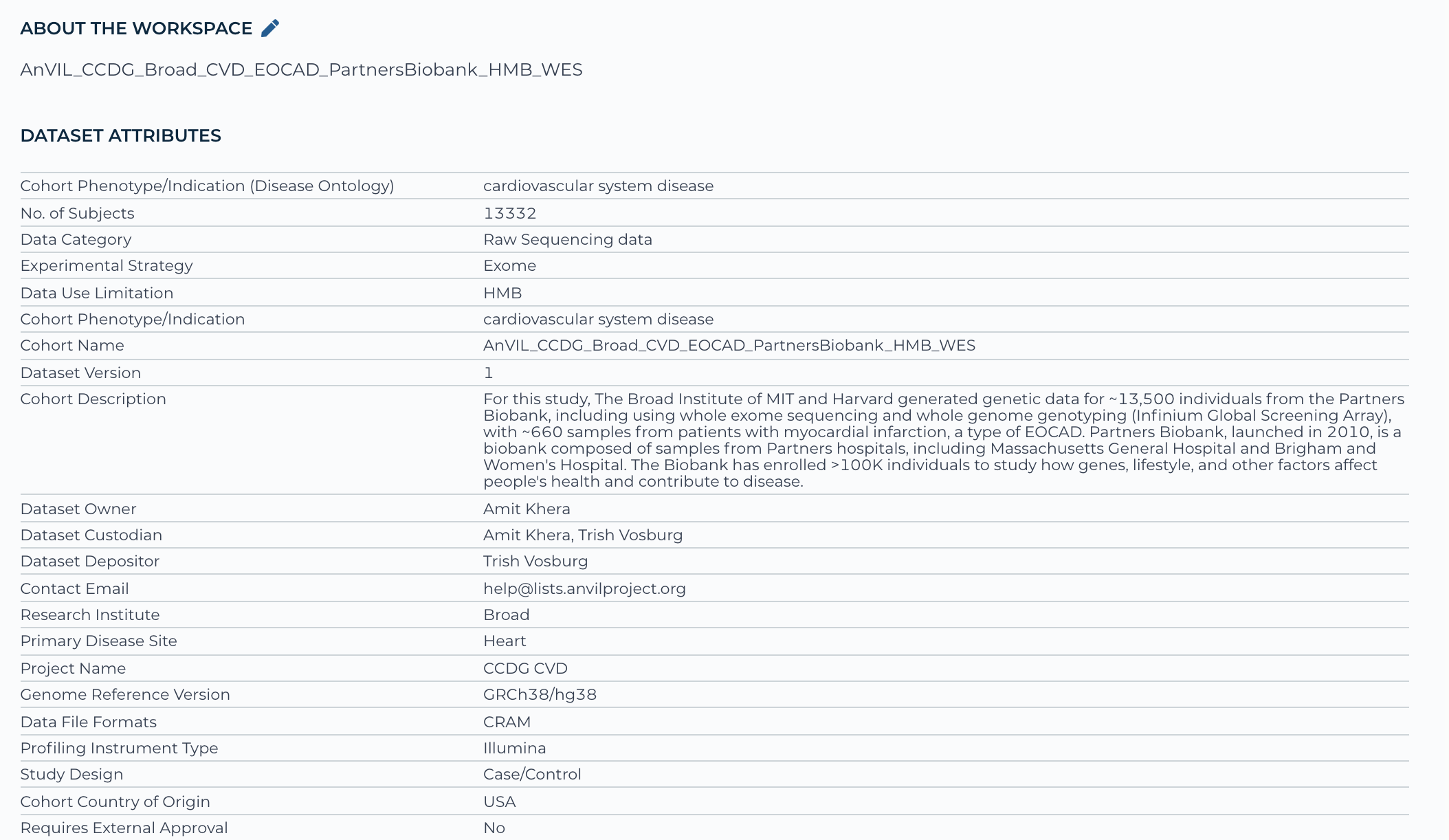
Example: AnVIL workspace - AnVIL_CCDG_Broad_CVD_EOCAD_PartnersBiobank_HMB_WES - represents 1 study registration phs002018 and has one consent group, which is health/medical/biomedical or shortened to HMB
Below is a screenshot of the data elements incorporated on the front (documentation) page of the workspace:
For Non-NHGRI funded studies that must seek Institutional and/or AnVIL Data Ingestion Committee approval (see steps 1.2 & 1.3), you may want to register your data in dbGaP while obtaining approval to speed up the administrative aspects.
Though there will be no requirement to submit source files or individual samples through the dbGaP portal, the dbGaP consent codes will be used to determine data access. Studies with multiple consent codes will be split into individual data workspaces based on cohort and consent pairings. External researchers can use dbGaP to apply for access, and a completed and approved DAR will permit dbGaP to link this access grant to Terra.
Data sharing
All individual-level human genomic and phenotypic data must conform to the NIH Genomic Data Sharing Policy. This includes the expectation that participants [are/were] explicitly consented for data sharing.
Access Control
For controlled-access datasets, access control within the AnVIL is governed by three major groups - developer access, consortium access, and external researcher access (via dbGaP). For more information, see Data Access Controls.
Getting Help
Please contact the AnVIL Outreach team with support and training requests at help@lists.anvilproject.org.